The Genome Asia 100K Project provides new data for medical and population genetics
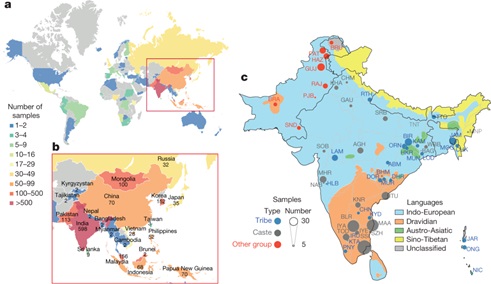
19.12.2019The underrepresentation of non-Europeans in human genetic studies so far has limited the diversity of individuals in genomic datasets and led to reduced medical relevance for a large proportion of the world’s population. Indian scientists launched the Genome Asia 100K project to solve this problem. They catalogued genetic variation, population structure, disease associations and founder effects for 1,739 individuals of 219 population groups and 64 countries across Asia.
The researchers used disease-related GAsP data to determine how the results of longer genomic studies in Asia can help to improve human health.
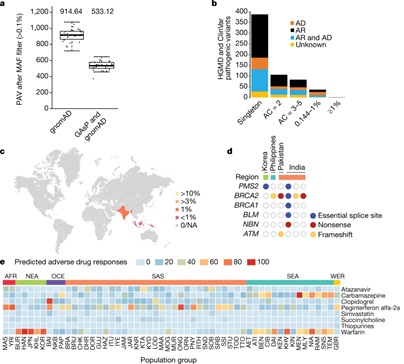
As expected the majority of coding variants were singletons or very rare. However, the absolute numbers of novel variants with a minor allele frequency (MAF) ≥ 0.1% within our pan-Asian dataset is large (n = 194,585), and these are frequent enough to be of relevance for large-scale genetic association studies. They also searched for variants present at low frequency in the overall dataset that are present at significantly higher allele frequencies in one or more of the population groups. The researchers found an additional 144,329 novel variants with MAF > 1% in the full GAsP dataset that were present at a frequency of greater than 1% within populations grouped by geography; South Asia, Southeast Asia, Northeast Asia or Oceania. These geographical regions contain many diverse population groups, and additional studies are needed to characterize patterns of genetic variation in these groups and disease relevance.
They also examined dataset for novel variants in genes known to be associated with cancer risk. We found 13 unique variants in 6 genes from 17 samples. This included frameshift, stop-gained and essential splice-site mutations in BRCA2 (n = 9), BRCA1 (n = 1), ATM (n = 2), BLM (n = 1), NBN (n = 2) and PMS2 (n = 2).
Variation in drug responses are generally recognized and recommendations for dosing are sometimes guided by apparent or self-reported population identity despite the lack of a rigorous pharmacogenomic basis. We assessed the allele frequencies of key pharmacogenomic variants in our dataset to identify inter-population differences that have potential implications on drug testing and treatment/
In additional, important data have been discovered that explain a slightly increased amount of Denisovan ancestry in south Asians compared with a priori expectations. They examined whether this was correlated with either language or social and/or caste status. South Asian samples were grouped into individuals who speak Indo-European languages and individuals who speak non-Indo-European languages, as well as four social or cultural groups: tribal (Adivasi) groups, lower-caste groups, high-caste groups and Pakistani groups (Indo-European language speaking only). They found that the average levels of Denisovan ancestry were significantly different between the four social or cultural groups. Their results are consistent with the scenario that Indo-European-speaking migrants who entered the subcontinent from the northwest admixed with an indigenous South Asian (ancestral south Indian) group who had higher levels of Denisovan ancestry.